
Cancer encompasses a broad spectrum of diseases (>100)
arising from the accumulation of somatically acquired mutations. These
mutations provide a growth advantage to a tumor cell, resulting in clonal
expansion leading to cancer progression. Among cancer-related mutations are
loss-of-function mutations in tumor suppressor genes and gain-of-function
mutations in oncogenes. Both tumor suppressor genes and oncogenes may be
regulated by a class of short (~21 nt long) single-stranded noncoding RNAs
known as microRNAs (miRNAs), that are intensively studied in cancer. They act
predominantly by binding to the target mRNA 3’ UTR sequence. Although miRNAs
are intensively studied in cancer and many miRNAs playing important roles in
cancer have been already identified, the cancer somatic mutations in miRNA
genes have been never studied before. It can be assumed that mutations in miRNA
or its precursor sequence may alter the folding of the miRNA precursor
structure, efficiency, and specificity of miRNA-biogenesis and the sequence of
the miRNA itself, thus altering the spectrum of target mRNAs. The somatic
mutations may both disrupt naturally occurring miRNAs and create new miRNAs
that recognize targets completely different from the original ones, which may
result in cancer development associated with gain- or loss-of-function of a
specific MIRNA gene.
Therefore, the main
objective of the project is to identify cancer-driving somatic mutations in miRNA and
miRNA-biogenesis genes.
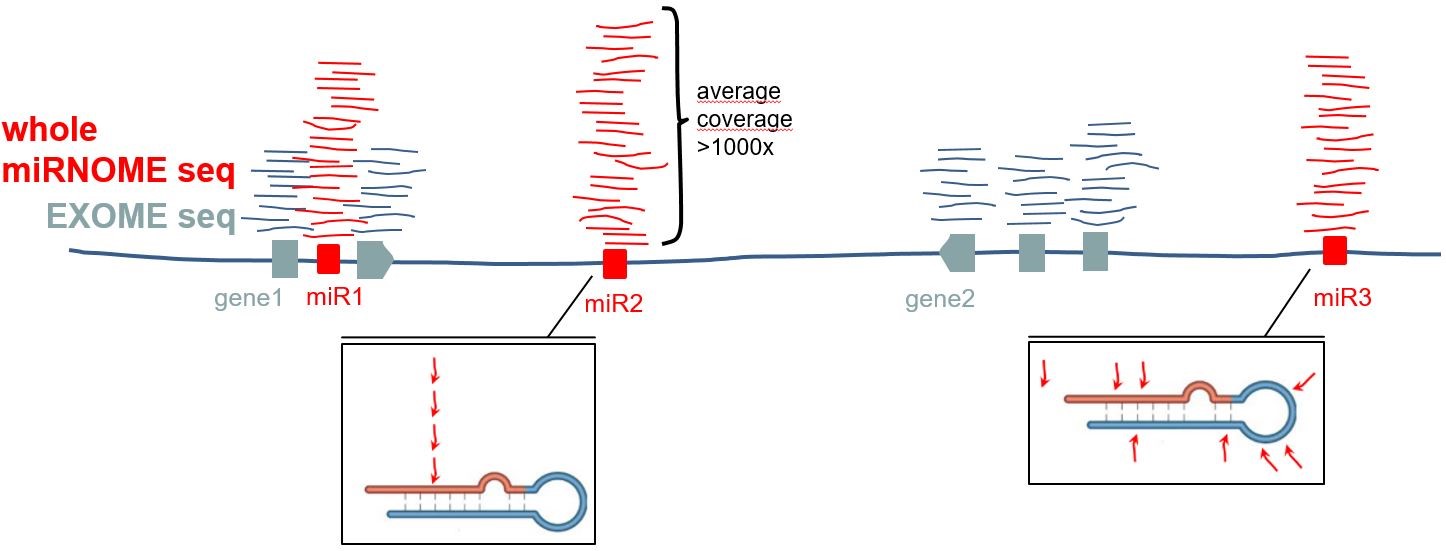
it’s hard to believe that at least
part of genetic variation driving cancer is not hidden in the non-coding
genome, especially in elements playing an important role in cancer, such as
miRNA genes
This website uses cookies to ensure you get the best experience on our website.
